Mechanisms of superinfection exclusion in archaea
Emine Rabia Sensevdi – Hector RCD Awardee Tessa Quax
Viruses are much more than parasites with a negative impact on the host. They can infect all domains of life and have different types of relationships with their host: From a parasitic to even beneficial relationship. One potentially beneficial relationship in favor of their host cell is the ability of some viruses to prevent superinfection by other viruses, which is known as superinfection exclusion (SIE). However, our understanding of this mechanism is rather scare. This project aims to decipher the molecular mechanism underlying SIE in haloarchaeal viruses using molecular and virological techniques.
The word `virus‘ is usually associated with a parasitic life form that negatively impacts the host cell during infection. However, viruses are far more than that. They are an essential part of life, capable of infecting members of all three domains of life. One domain of particular interest are the micro-organisms belonging to the domain of Archaea, since little is known about their interplay between virus and host. Archaea and their viruses represent a significant part of microbial diversity on earth. It has been suggested that in some environments dominated by archaea, half of the cells are infected by a virus at any given time point. It is therefore very likely that a new virus will encounter an already infected cell, which would lead to competition for cellular resources. To circumvent this, some viruses have evolved the ability to block infection of their host cell against superinfection by the same or another closely related virus. This phenomenon is known as superinfection exclusion (SIE). However, knowledge on superinfection exclusion is still rather scarce, and the molecular mechanisms behind SIE have mainly been studied for viruses infecting bacteria. Such mechanisms still have to be unraveled for archaeal viruses. This project aims to provide a first insight into the requirements for superinfection exclusion in archaea, and would therefore provide essential information to further investigation of the molecular mechanisms underlying this intriguing phenomenon in haloarchaeal viruses. To achieve this aim, I will use a combination of qRT-PCR, fluorescence and electron microscopy.
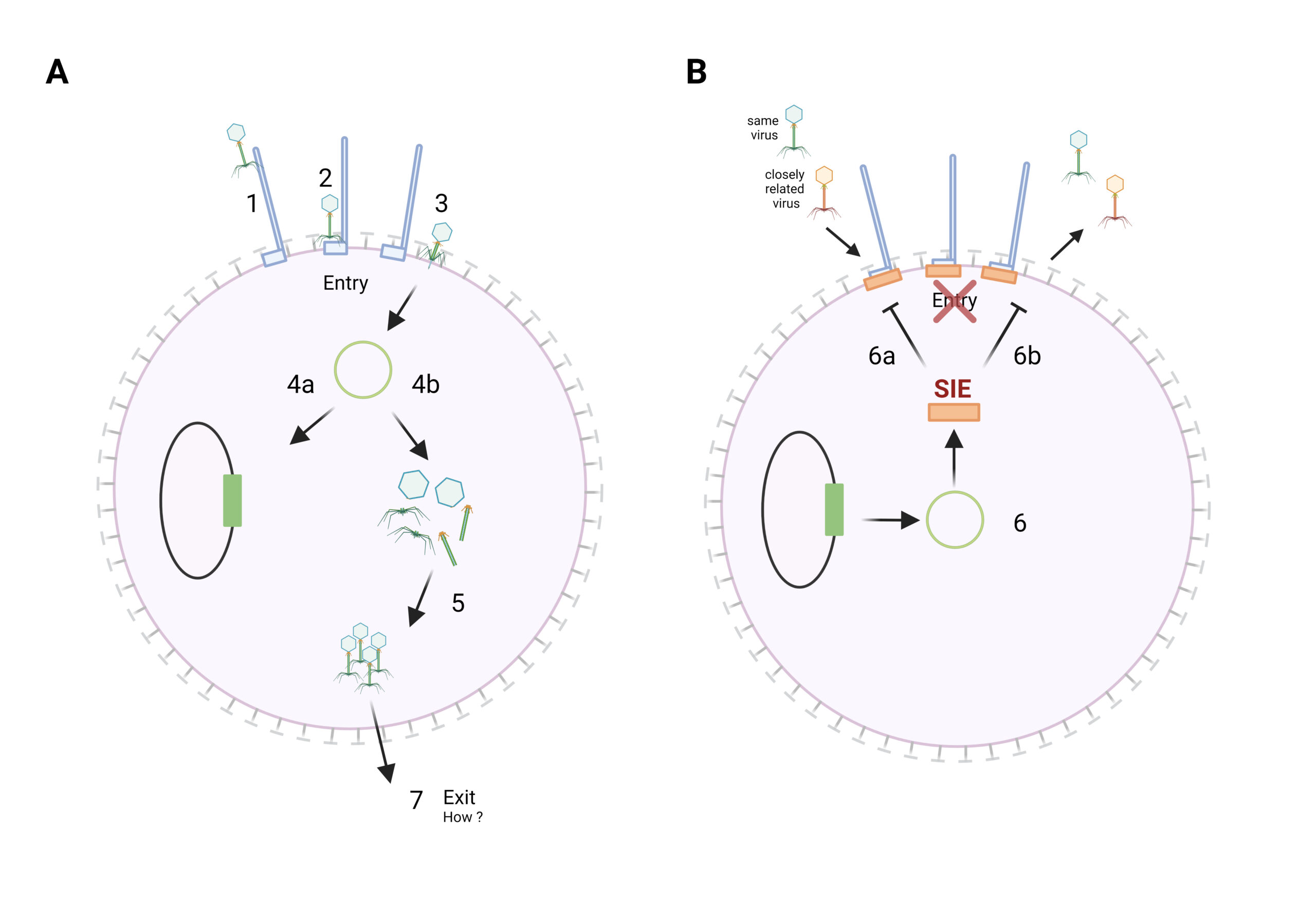
Superinfection: (A) Succesfull infection with a virus (green). Virus attach either to surface filaments (1), travel to cell surface (2), or passage their genome over the cell membrane (3). The viral genome can be integrated into the host genome, also called lysogeny (4a), or replicate and form new virions (4b and 5), which are then released from the cell, e.g by cell lysis (7). (B) Inhibition of Superinfection: strategy of viruses to inhibit superinfection by other viruses. The integrated virus may induce a genetic programme, that results in the inhibiton of superinfection of similar (green) or different (orange) virions (6) for example by blocking of phage-receptor (6a) or by alteration of cell surface (6b).

Emine Rabia Sensevdi
Rijksuniversiteit GroningenSupervised by

Hector RCD Awardee Jun.-Prof. Dr.
Tessa Quax
Biology
